180x Sensitivity Improvement in Cardiac Troponin I ELISA
180x Sensitivity Improvement in Cardiac Troponin I ELISA
Watch the Webinar Replay to See How We Achieved 0.07 pg/mL Detection
Enter your email to join Dr. Peter Stenlund as he reveals BOLD’s 50x sensitivity boost!
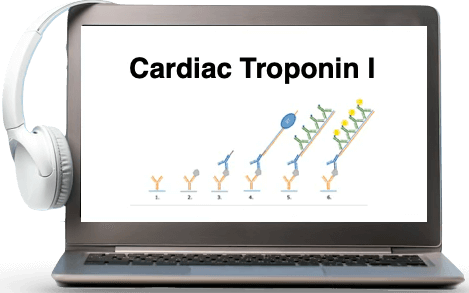
Good afternoon and good morning. Thank you for joining us today for our webinar, where my colleague, Dr. Peter Stenlund, principal scientist at Cavidi, will present a case study on our work developing an ultrasensitive ELISA for cardiac troponin. My name is Jonathan Royce, and I am the CEO of Cavidi. Today, I will be the moderator for this event. In just a moment, I will be handing over the presentation to Peter. Before I do, just a few housekeeping items. At the end of the presentation, there will be a live q and a session. You can, at any time during the presentation, write your questions in the q and a panel to the right of the presentation window. We will answer as many of your questions as we can at the end of the talk. If you experience technical difficulties, please also use the q and a panel to report the issue, and one of our producers will do their best to help you. Also, if you'd like to view the presentation in full screen mode, there is a small button in the lower right of the window which enables this mode. Now I am pleased to introduce Peter and hand the presentation over to him. Thank you very much, Jonathan. So hello, everyone. So I will, in this webinar, introduce you to single amplification technology that we have invented at Cavidi. It's called BOLD, and it can be used to enhance the sensitivity of immunoassays. And I will show you how BOLD works step by step. I will also show you how you can integrate it into an existing, immunoassay, and I will also show you how we improve the sensitivity of a cardiac troponin eye sandwich ELISA. But before we start with that, I'd like to emphasize the importance of ultrasensitive immunoassays and why they matter in life sciences. I have highlighted three areas here, drug discovery, biomarker discovery, and detection, which is a more general area. And also, I have highlighted early disease detection where it may, matter a lot. So if you look at drug discovery, well, of course, you might improve identification of potential drug targets, of course, having an, ultrasensitive immunoassay. It can also deepen the understanding of disease mechanism, and you might, improve, or you can improve the pharmacokinetic study where you can measure low drug levels and also better understand biological effects. And, of course, in clinical trials, you can early you can detect early disease markers, for example, that may help you understand how how your drug is working from a safety and efficacy perspective. In the more general area, you have, of course, the possibility to detect and discover low level biomarkers. And also when you are working with small animals, the volumes might be scarce, so that will help you to make more use of more use of your small sample volumes. And in early disease detection, for example, you will have the possibility to detect disease biomarkers at at very low levels. And these are especially important in neurodegenerative diseases where you can detect very early and all also before you can see, for example, cognitive decline in Alzheimer's disease, for example, and also early detection in recurring cancer. And, of course, typically, you measure biomarkers from Alzheimer, for example, by taking samples from cerebral spinal fluid, it might be possible to detect these biomarkers in blood with an ultra sensitive method. So and also, this is, of course, crucial if you would like to sort of intervene early before the disease have have sort of manifested. So with that, I'd like to sort of emphasize the importance of ultrasensitive amino acids and the benefit. So let's go into BOLD. As I mentioned, BOLD is short for binding oligo ladder detection, and it's a signal amplification technology that you can use to amplify the signals in immunoassays. And I have listed here some benefits of this, technology. And the the first one is you can see up to a two hundred fold improved sensitivity through the signal amplification, and the amplification is an molecular biology approach that I will show you in more detail. And I also listed its cost effective add on technique, so you add it to your existing immunoassay. There is no need to invest in expensive new equipment. You use the instrument that you are using for your immunoassay, if it's an ELISA or another type of technology. You can also use your previously optimized and validated antibody pairs. And there is a minimal extension of acid time, I'd like to say ten to sixty minutes in a standard ELISA. And there is really no limitations what kind of biomarkers that you can detect. And another positive thing is that it's easy to, it's easy to let's see here. If I can get the the laser pen. Sorry. Yeah. So it's easy to to implement in existing immunoassays. So this is an a sandwich immunoassay. It could be an ELISA, for example, or it could be on a a bead of some sort. So we have a capture antibody. We have an antigen, and then you have a detector antibody that typically is biotinylated. So you can, for example, use streptavidin conjugates for detection. And the typical conjugates are like HRP, for example, or alkaline phosphatases. And this is then ELISAs, and you can also have streptavidin conjugated to fluorophores as or or, like, protein molecules such as hypoerythrin and and APC also as well. But this is the standard immunoassay. If you want to implement both, you need an unmodified detector antibody. And to this antibody, you conjugate an oligo dt primer that is around thirty base pair sorry, thirty base bases long. And this oligo dt primaries conjugated to the antibody through click chemistry. So you use the amine groups on your detector antibody, and you use click chemistry. So the first step is to modify the detector with an a side molecule. And this is done using a click label and with an a side type tag. It's an an NHS ester. And then, you remove unreacted esters, and, you go to step two where you add, an alkyne, which, a a dBCO modified oligo dt primer. And this dBCO moiety reacts with the a side group specifically, and you get your conjugated detector. And once you have this detector, you can continue with BOLD, and you add poly RA template, which is around two hundred base pairs long. And in the next step, you add a DNA polymerase with reverse transcriptase activity, and you also add some, brominated nucleotide triphosphates, BRD UTPs. And this polymerase builds up a DNA RNA hybrid, and this takes around ten to sixty minutes depending on on how long time you'd like to or how much of amplification you you need. So then you have a DNA RNA hybrid with BRDU, and we add tertiary biotinylated anti DNA RNA antibody that recognizes this BRDU, nucleotides on the DNA RNA hybrid. And then you can add your streptavidin conjugate for detection. And as with the standard ELISA, you can use the same streptavidin conjugates for detection. So let's start to develop an ultrasensitive human cardiac troponin eye sandwich, ELISA. So, before we do that, I just like to talk a little bit about cardiac troponin eye. Troponins are are proteins that regulate skeletal and cardiac muscle contraction. That's sort of the the the basis of troponins. And troponins, they form a heterotrimeric complex that contains troponin c, troponin I, which we will talk about today. And this troponin I inhibits actin myosin interaction, and you also have TNT in your complex. And cardiac troponin I, for example, is the gold standard for gold standard biomarker for myocardial infarction for cardiac injury. And typically, after a cardiac injury, car cardiac troponin appears in the bloodstream, and you can detect it with diagnostic immunoassays. And there are some particular concerns here because there also exists skeletal skeletal troponin, which can cause cross reactivity. So this means that you need really specific antibodies in your immunoassays that only recognize cardiac the cardiac form. So if we look at normal levels, in men, for example, we have twenty picograms per mil or less. And in women, we have a little bit lower, which is related to that there is a variation in sex and also variation in age and kidney function and renal function. And also some diagnostic clues here is that if you have an increase, a two to threefold increase, that may indicate some sort of cardiac injury. And it's also, as you might understand, important to be able to detect cardiac release of cardiac troponin at a very, very early stage. And that's why these high sensitivity assays are so important. And it also may help to rule out, occasions where there is no real cardiac injury in the patients. And also, of course, it's important to understand some clinical cardiac conditions as well to to be able to measure really normal low levels with great precision and accuracy. So in this webinar and in this development, I will walk you through how we selected capture and detector antibodies. I will also talk about how we evaluated capture antibodies for the standard and amplified assay. It's it's a comparison, more or less, a benchmarking. And also, I will show you how we evaluated detector antibodies for standard and amplified ELISA. And, we use some different ways of conjugating. And, also, I will talk a little bit about downstream assay optimization where we sort of tweaked the the last points of the development to to get as good assay as possible. And we also did some performance testing of standard and amplified ELISA. We did that in room temperature, in in a simple buffer and also in serum at room temperature, and also we tested it in initial test at thirty seven degrees in some of these automatic instruments that measure cardiac troponin works at thirty seven degrees Celsius. So the first step then, selection of detector and capture antibodies. Here I have amino acid sequence alignment, and, I have aligned cardiac troponin, which is the the top row, and then I have, the slow and fast skeleton, troponin eye. And as you see, the first thing is, of course, you see that there is no counterpart in the skeletal variants of this region that only the cauda troponin has. And if we look at everything that is red, that means that we have one hundred percent identity. We have the same amino acids at at those positions. And then if it's blue or black, we have some some differences. So if you look at certain regions here, you'll find the areas that is best well, what that has the the biggest difference. And I have highlighted them here. So it's the n terminal, and you see an area between eighty and hundred and also between a hundred and sixty and two hundred in this sequence stretch. And we selected, not by random, but we selected a couple of antibodies that we wanted to try. The first one was VEK R33, which was in the beginning of the troponin sequence, and we also selected nineteen c seven c c, which is a bit downstream. And we also selected five six o cc and sixteen a l a eleven cc. And, also, finally, we choose six hundred and twenty seven. This is the clone names, and all of these were purchased from HITEST, which is a Finnish company. And they have more than fifty different variants of antibodies, clones for antibodies against the cardiac troponin I. So the first thing we did was to evaluate capture antibodies for the standard and amplified ELISA. So here, I have four figures. I show you absorbance values in the top row. And we looked at three antibodies, and these are sort of single tests where we have a surface only with one clone. And to the right, we have two of the clones at the same time. And we tested at one micrograms per mil during the the capture step and between o point sorry, and o point five for each of the pairs simultaneously. So if we look here, we see that all of them have almost the same sort of signals. Nineteen c seven c c perhaps is a little bit worse. If we look at them together, we really see no big difference here. We have the same scales here. And if we look at the signal to noise ratio, we can see that that we I mean, that they all have sort of comparable backgrounds across all these antibodies. And as I said, six twenty five and RecR thirty three compared similarly well, and they all have additive signals when they were combined. And one good thing with having dual antibodies that is that they have different specific specificities for cardiac troponin isoforms. So depending on, let's say, if you have if if it's in the in a complex, for example, or if it is proteolytically cleaved in some way, it's good to have more than one epitope that you use to to detect a cardiac troponin I. So in the next step, we did the same for the amplified ELISA. And as you see here, we have much stronger signals. It's up to around three compared to around zero point five for the standard ELISA. We did the same setup here. We tested one, and we also tested two simultaneously. And if we look at absorbances here, we have greater variations in the absorbent signals among the antibodies compared to the standard ELISA. There is some sort of difference in background signals across all of the antibodies. And we can see that if we look at these where we only have one, we go down to the signal to noise ratio. We can see that, for example, six to five is starting to perform better at the same yeah. It's starting to perform better here. So there is something with the background that that is different between these antibodies. And when we are working with signal amplification background, it's very important to keep as low as possible. And then if we look at using them both at the same time, we see a greater change in absorbance with combined use of antibodies compared to standard ELISA. And if we look at the signal to noise ratio here, we see that they start to sort of they're pretty much similar despite this difference in raw signal. And then if we look at yeah. And and the same also, the same is important even in the amplified ELISA that it's good to have more than one, of course, antibody to to capture the antigen. So there is something with the background. So if we take a look at the background then, we can see here in the figure to the left where the orange are the amplified ELISA, and this is the background signal for each of the clones that we tested. And we clearly see that for, ZR thirty three, we see a significant higher background for some reason. And this is something that we really don't want when it comes to signal amplification. So we ended up we wanted to have two at least two antibodies as capturing antibodies. So we ended up using nineteen c seven cc and also six two five. So we used them in all of the upcoming experiments. And we also looked at detection antibodies, but first, I just like to show you that we used two types of conjugation processes. The first one I showed you in when we talked about bold earlier, you modify your antibody with an a side, and then you go in the next step and modify it with dBCO. So you get to conjugate, and you and in conjugation process two, you do almost the same thing, but you do the reversed thing here. So you modify the antibody with dBCO in the first step at amine groups. And then in the next step, you use an A side modified oligo dt primer. So it's really the same but reversed. And, here are two figures showing, where we evaluated the detector signals, the absorbances. And for, the standard ELISA, we only, we were limited to use only two detectors. So we used sixteen a eleven cc and five sixty cc since we only have had them biotinylated. And for the amplified ELISA, we used the same two antibody clones, but also these RecR33. So we tested in the standardized, we tested at one micrograms per mil, which is typical for a standard ELISA. And we could see clearly there that we had better signals from, using sixteen a eleven, And it wasn't as clear when we look here at the amplified ELISA. We see a lot of higher signals, of course. But if you look here, the the closed circle here, we use a process one for conjugation of five sixty cc. And in the open circle that we see, we used process two, the reversed variant. We tested all of these detector antibodies at the same concentration. And when we look here, we really see nothing that sort of stands out, really. We noticed when we did different ratios of the first step when we modify the antibody with the click m label and the next the second step when we modify it with the primer. So twenty five, that means that we used a twenty fold molar excess of the ClickCAM label, the first step. And five means that we use a five fold excess compared to that ClickCam labeled antibody in the second step. So this only shows the performance of this ratio, and that performed best. We tested higher ratios as well and also lower ratios of click and label and primer, but twenty five works the best. But here, it's difficult to see a really clear clear best performer, but if we look at the signal to noise. Also, one thing to mention is that we are looking here at o two sixteen picograms per mil on amplified ELISA, and we are looking at two points at a hundred and one thousand picograms per mil for the standard assay. And it's really this shows a really strong signal amplification. So if you look at signal to noise ratios, then the standardizer looks pretty much the same. There is no change, and that's because there is no real difference in background between the two. It's still the best performer. And if we look at the amplifying analyzer, we can see that there is something that sort of pans out here We see that the the highest signal to noise ratio we obtained using five sixty cc using process two for conjugation. And if we compare it to process one, we see that using process one results in a significant increase in in background. And in as when we we are working with signal amplification, we really don't like background problems from the beginning. So we picked out five sixty cc using process two for conjugation. So we went further further and tested the effect of concentration on this detector for the amplified ELISA. We didn't do this for the standard ELISA because the the concentration that we use, one micrograms per mil, worked perfect. We didn't see any improvement if we used a higher concentration or a really lower concentration. So here we see different concentrations of the detector antibody, and we see that we have the the lowest thing that we obtained from the lowest concentration, of course. And if we go up to zero point one micrograms per mil, which is onetenth compared to the standard ELISA, we see a significant improvement. But if we go from o point one to o point two, we didn't see a really good improvement. And also if we look at signal to noise ratios, we can see that all of them almost end up on the same sort of signal to noise ratio. So in fact, we in this case, we we selected zero point one micrograms per milliliter because we didn't see that much of an effect compared to using twice the twice the concentration. And also, we see that we have a lower background compared to when we use o point two micrograms per mil. So we selected to run all the following experiments with o point one micrograms per mil. And we also did some additional parameters that we we evaluated. And I I don't show any results here, but it was related to coating, for example, where we looked at temperature and time, and we found that overnight was best. It was also convenient, but at a concentration of two to eight degrees Celsius during incubation. We looked at blocking solution, and we tested parameters such as PBST, which is phosphate buff buffer saline with some tween, o point o five percent tween. And we also tested additives such as casine, NFDM, and also BSA. And we ended up with a blocking solution with PBST and one percent of caffeine. And we also looked at sample and calibrator dilution, and we looked at the same type of buffers for convenience also. And we ended up using o point one percent caffeine added to PBST. We also looked at polymerization, and that is only for the amplified ELISA, of course. We looked at polymerase concentration and also polymerase reaction time. And we found the best conditions were obtained using a concentration of one point three units per mil of the polymerase and the extension time sixty minutes in this particular case. And the last step in a bold assay is the detection using the tertiary anti DNA RNA antibody, and we looked at concentration. We evaluated the concentration of the antibody and also the time of incubation, and we ended up using o point three micrograms per mil. And the binding or the incubation time was thirty minutes. So these steps are sort of specific for both, but all the other steps were also tested for the standard assay. So we tested some performance, and we compared the standard and amplified ELISA. And in this figure, the orange is the amplified ELISA, and the black is the standard ELISA. And we are looking at the concentration ratio of o to sixteen picograms per mil for the amplified ELISA and between o and four hundred and thirty two picograms per mil for the standard ELISA. And as you can see, there is a real significant signal amplification that is achieved using BOLD. And where the standard assay all only shows background, we really see a significant signal from from the the ELISA using BOLT. And also here, just to let you know about precision, for example, and this is repeatability. And this column is for the standard ELISA, which shows pretty low CVs CV percentages between two to five. And if we look at the amplified ELISA, we can see that it's also very low acceptable. And we can look at, for example, the first calibrated point for the the amplified ELISA. And if we go down to this black point here for the standard ELISA, we see that we have a small advantage compared to the standard ELISA. But, anyway, it will not be possible to quantify any cardiac troponin I in this point in the standard ELISA, whereas it's very doable using the amplified variant. So this also was in a simple buffer. So it's it it gave sort of the the method the best opportunity, but we also tested it in twenty five percent serum. And we had the same sort of calibrator ranges. And this is an average of three independent runs. So these are the signals that was obtained in absorbance for three different runs. And we wanted to see the LOD here, so we wanted to estimate that. And, typically, you estimate LOD by adding two standard deviations to your background, and then you go into your standard curve and you get the concentration. But in this case, we chose to look at LOD or the signal at two times the background. So we really have a signal that we can can compare. And by doing that, we found that for the amplified ELISA, we had an LOD of o point o seven picograms per mil. And for the standardized, we had an LOD of twelve point five picograms per mil. And twelve point five is pretty common for commercially available ELISAs out there. And if we compare these two, we can see that we have almost a two hundred fold improvement, while using, an amplified ELISA compared to the standard ELISA. So we were very happy to see this. And, also, one thing with detecting cardiac troponin is that many of these automatic instrument, they use, that they detect cardiac troponin at thirty seven degrees Celsius. So we just wanted to see if that would be possible and also possible to see if we would have a signal, enhanced sensitivity in in the temperature. And the only thing is that we needed to use a different polymerase because the polymerase that we use at room temperature does not work well at thirty seven degrees. So we we took another polymerase also with reverse transcriptase activity. And we did the same testing here to derive the LOD. We used a signal to background ratio of two. And as you can see here, just by looking at the calibration curves, we see that we have a significant signal amplification still. Although we have a little bit higher LED for the amplified ELISA around sevenfold, and we have a twofold difference. Yeah. Almost twofold difference compared to the the when running the standard ELISA at room temperature. But still, we can see a fifty fold improvement, which is really good. So to conclude this presentation, so at room temperatures in serum, we obtained an an a hundred and eighty fold increase in set sensitivity using both compared to the standard ELISA. And we also saw, as I mentioned just before, that it's possible to to also have signal amplification at elevated temperatures such as thirty seven degrees, which is common for automated systems. Although we saw a little bit lower improvement, but we haven't we but they didn't sort of optimize the method for that temperature. I also think that it is important to mention that it's important to test different combinations of capture and detection antibodies And especially for cardiac troponins, this is important, and and also other biomarkers as well. And we also saw that systematic optimization of other assay parameters and capture antibodies and detector antibodies were very important. We did some changes with reagent concentrations, incubation times, and also looked at enzymatic activity of the polymerase. And this was important to maximizing the performance of the BOLD assay. And we didn't observe a clear boost in sensitivity by simultaneous use of different capture antibodies. But we expect a better performance when this assay is used for detecting partially degraded forms of cardiac troponin and also when a cardiac troponin eye is within the complex. And I also like to mention that here we only looked at cardiac troponin eye as a monomer, So we didn't do any testing on on it didn't show any testing on the complex or how, for example, the skeletal skeletal variants might cross react. And also future optimizations, of course, evaluate the current sort of antibody pairs and also look into other antibody pairs that may be better at detecting degraded forms of cardiac troponins and also when cardiac troponin is within a complex. And, also, I know that the high test have humanized antibodies that is good when you like to get into control over, heterophilic antibodies that may give you problems with false positives and false negatives. So I also like to acknowledge my colleagues here at Cavidi that has been involved in this project and done many of these experiments. That's Mariam, Jessica, Veronica, Johanna, and also Jonathan that's here today. And by this, I hope that I have given you some insight in how you can use BOLT to amplify your cardiac troponin assay or any other type of assay. And if you are interested in trying this technology out, you can contact me or Jonathan, or you can also contact us through our web page. And we I'd also like to mention that we have distributors in, of course, your region in Asia, Europe, and America. Thank you. Thank you, Peter, for an excellent presentation. We'll now jump into the q and a. But before we do, Cavidi has an exclusive offer for webinar attendees that I want to briefly describe. I know that many of you have assays that need better sensitivity, but you don't have the time or resources to adapt the method with BOLD. Therefore, we're offering to do the work for you free of charge. You have an assay which would benefit from BOLD, bottom the button at the bottom of the screen and fill out the application. We will be selecting up to three projects for this offer by November thirtieth. Now let's move on to questions. There are several here. So first, the dynamic range of the amplified ELISA seems to be quite narrow, being just between zero and sixteen picograms per milliliter it showed. Is that a limitation that comes from the ELISA and the absorbance measurement, or could the measuring range be expected to be wider with some other detection systems? Yeah. That's a that's a good question. Yeah. I mean, this is absorbance, and, typically, you don't get that many magnitudes of dynamic range. It's it's very, highly sort of amplified. We have seen, for example, that using other detection modalities such as luminescence, for example, we have seen up to almost four magnitudes of dynamic range. So I think it's more well, it's two things. It's the the the power of amplification, and it's the detection modality, I guess, is responsible for this. Okay. Thank you. Another question that we had here was, when we were screening for the capture antibody, how was the analyte detected? So what was the was there a I think the question is, was there a standard detection antibody that was used during the screening of the capture antibodies? Yeah. I mean, it's like the hand and the egg. Where where do we start? So, we were limited to use two detector antibodies for the standard assay, and that was sixteen l eleven a c c and and five sixty cc. So for that purpose, we used sixteen eleven sixteen a eleven cc for the standard ELISA. And for the the amplified ELISA, we use five hundred sixty cc. Yeah. That's that's the answer. Yeah. Very good. Another question is, can you give a sort of general guideline about what sort of signal amplification a user should expect when using Streptavid and HRP? What is the sort of normal magnitude of amplification that we expect for a given analyte concentration? Yeah. I mean, that that if you look at the calibration curve, you will see that that the amplification sort of varies over the range. So it's not like five times all over the all over the the sort of calibration curve. So it's very difficult to say. I mean, if you compare, like, a fluorophore to a HRP, for example, you will have signal amplification with a HRP. So HRP by itself is a signal amplification variant. So it's I I I have difficulties in answering that question. It's a bit case by case. Yeah. I would say that. Oh, okay. We have a question here. Can Bold be adapted for multiplexing? Yeah. I mean, if yeah. I understand the question. So, typically, if you look at multiplexing, in in many cases, you use beads, and many of these beads have identities. So if if you sort of know the the biomarker if you sort of decide what biomarker each bead should have, it shouldn't be a problem using BOLD, of course. You will need you will have the same detection modality using the touch area antibody that that Bold use. So the bead will sort of differ between all the biomarkers, and Bold would do the same work on all beads. Okay. Perfect. I think that covers our questions. And, in fact, that is all the time we have for today. In about two weeks' time, you will be receiving a link to a recording of the webinar in case you wanna return to the material. Thank you again for joining us, and goodbye.
What You’ll Learn in This Webinar Recording
- How BOLD technology supercharges standard immunoassays
- Case study: Achieving a 180x lower limit of detection for cardiac troponin I
- Step-by-step guide to integrating BOLD into your existing ELISA workflows
- Future applications for drug development and clinical diagnostics
Peter walks you through optimizing a human cardiac troponin I ELISA, achieving a limit of detection (LOD) of 0.07 pg/mL—nearly 180x better than the standard 12.5 pg/mL. This breakthrough not only enables earlier cardiac injury detection but also provides the sensitivity needed for rapid clinical decision-making and improved patient outcomes.
About Peter Stenlund
Dr. Peter Stenlund is an experienced protein chemist with a PhD in biochemistry from Umeå university. Briefly, from that point forward to his present position at Cavidi AB, he has been employed as a senior lecturer, as a post-doctoral scientist in USA, as a development scientist at Biacore and as a senior scientist at Octapharma and Galderma. His work has focused on analytical development aimed for characterization and release of protein drugs.


About Exazym®
Exazym® is an add-on immunoassay reagent kit that provides attomole-level detection of low-abundance biomarkers using standard immunoassay workflows. It is based on a new detection method called Binding Oligo Ladder Detection, or BOLD for short.